The term “knowledge graph” describes the process of storing data in a graph-structured topology. A graph database consists of three main elements: nodes, edges, and properties. Nodes symbolize instances or entities, such as proteins, diseases, or tissues. Edges, otherwise known as relationships, provide a contextual connection between nodes. Properties, on the other hand, enrich both nodes and relationships with various data types such as texts, numbers, boolean values, or more complex structures like lists and maps.
History of Graph Databases
The concept of graph databases was first articulated by Austrian linguist Edgar W. Schneider in 1972 to discuss the creation of a modular interface for data analysis purposes. Initial attempts to create such a modular data analysis interface trace back to the 1960s. Today, graph databases underpin numerous technologies, from social networks, search engines, and maps to diversified Internet of Things (IoT) applications. They showcase particular efficiency in handling highly interconnected data and ensuring schema flexibility.
Current Adoption and Relevance in Bioinformatics
Despite their efficiency, graph databases haven’t gathered significant popularity, as reflected in the marginal number of PubMed database entries when searching for the term “graph database”, which only yields ~2% of the results compared to searching for the term “relational database”.
At Delta4, we advocate for the pertinence of graph databases in digital drug discovery and biological data management. Why?
Aside from technical implications, the dynamic adaptability of graph databases makes them a perfect fit for the fast-evolving, dynamic field of bioinformatics. Now, let’s contrast them with their conventional counterparts – Relational Database Management Systems (RDBMS).
Graph Databases vs. Relational Databases
Relational Database Management Systems (RDBMS)
An RDBMS forms an interconnected matrix of tables based on distinct table column values. However, in many circumstances, even minor modifications to tables at the foundational level of an application necessitate corresponding adjustments across various other layers. Despite its strict schema policy guaranteeing data consistency and strong mathematical operation capabilities, the RDBMS’s fixed structure can pose a hindrance when dealing with highly interconnected or frequently changing data types.
Graph Databases
Graph databases, on the other hand, offer simplicity and readability in representing complex relationships. For instance, a relational database schema accommodating molecular features, drugs, and phenotypes can be represented more intuitively in a graph structure, encoding details of the table into edges and properties.
Querying Data: Graph Databases vs. RDBMS
For illustration, let’s consider an example of a relational database schema accommodating molecular features, drugs, and phenotypes, as well as their interactions – a standard use case in digital drug discovery:
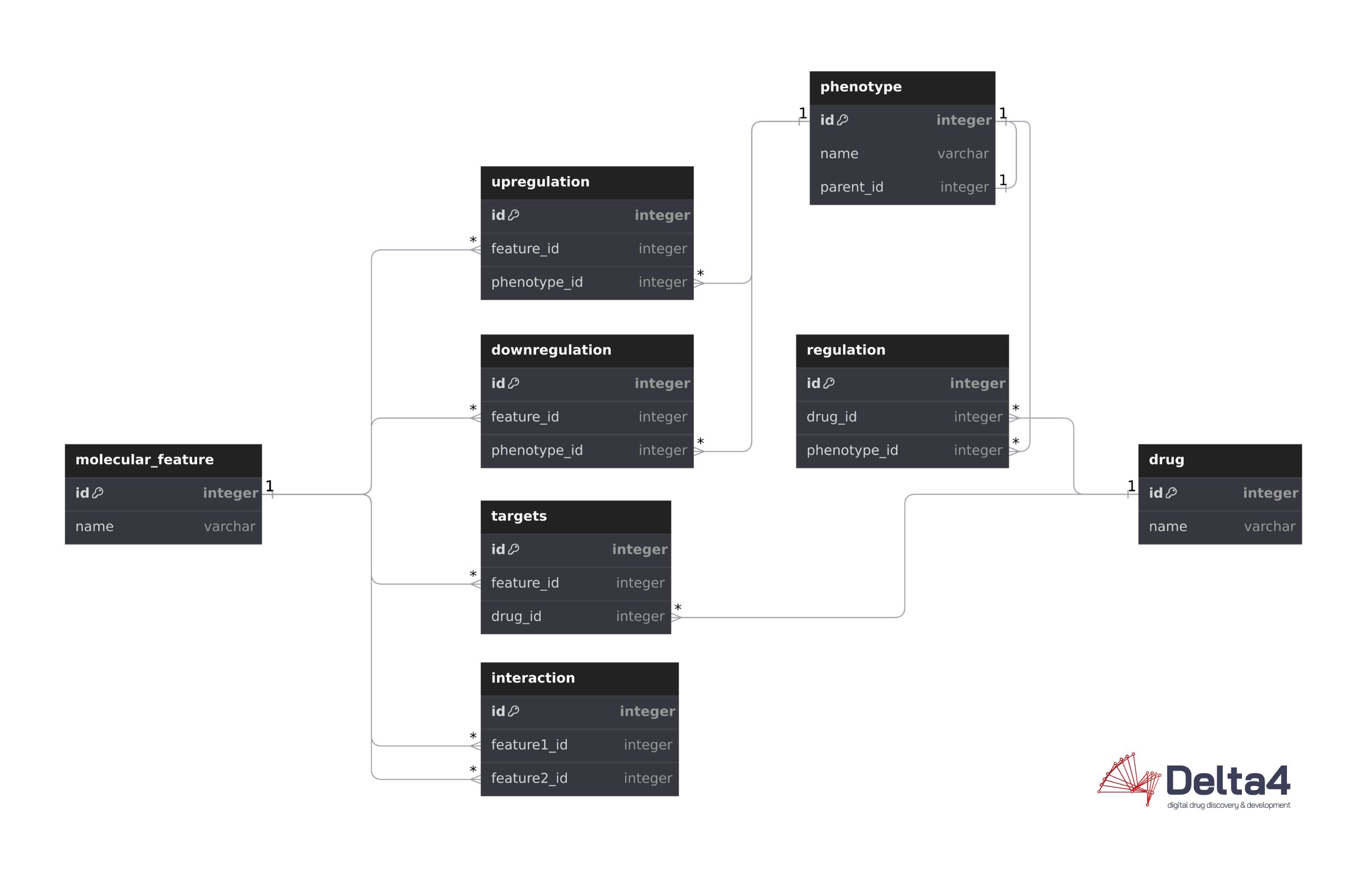
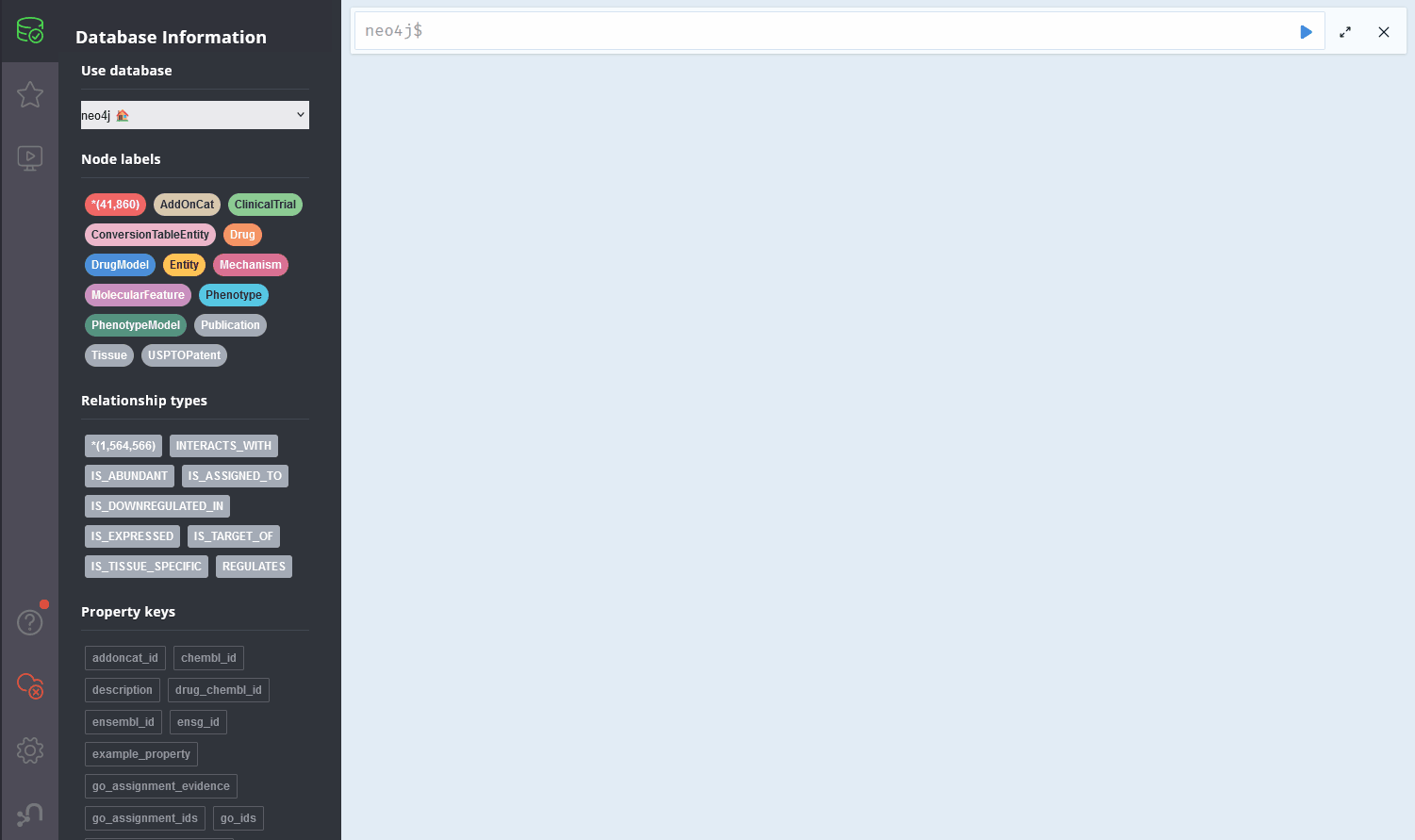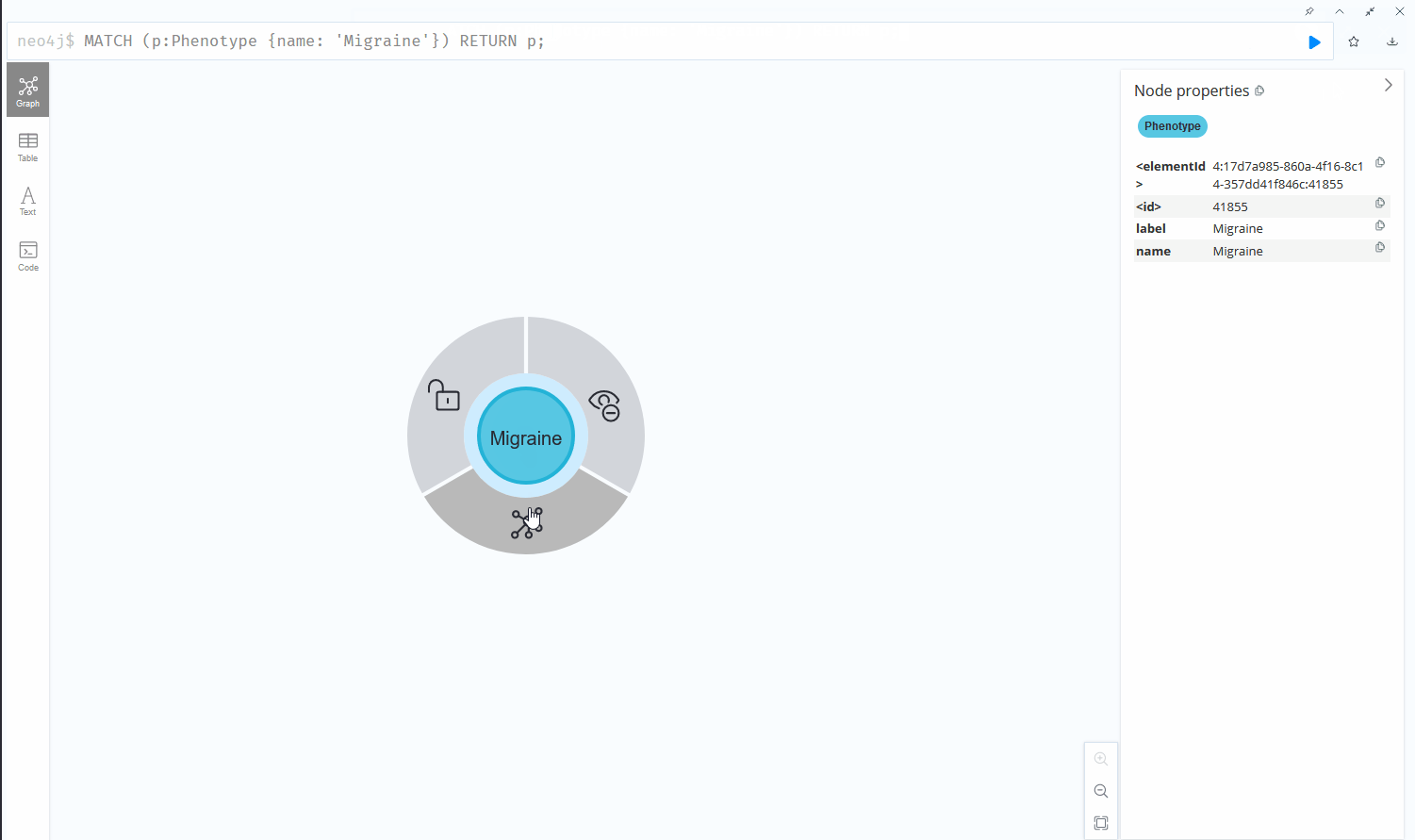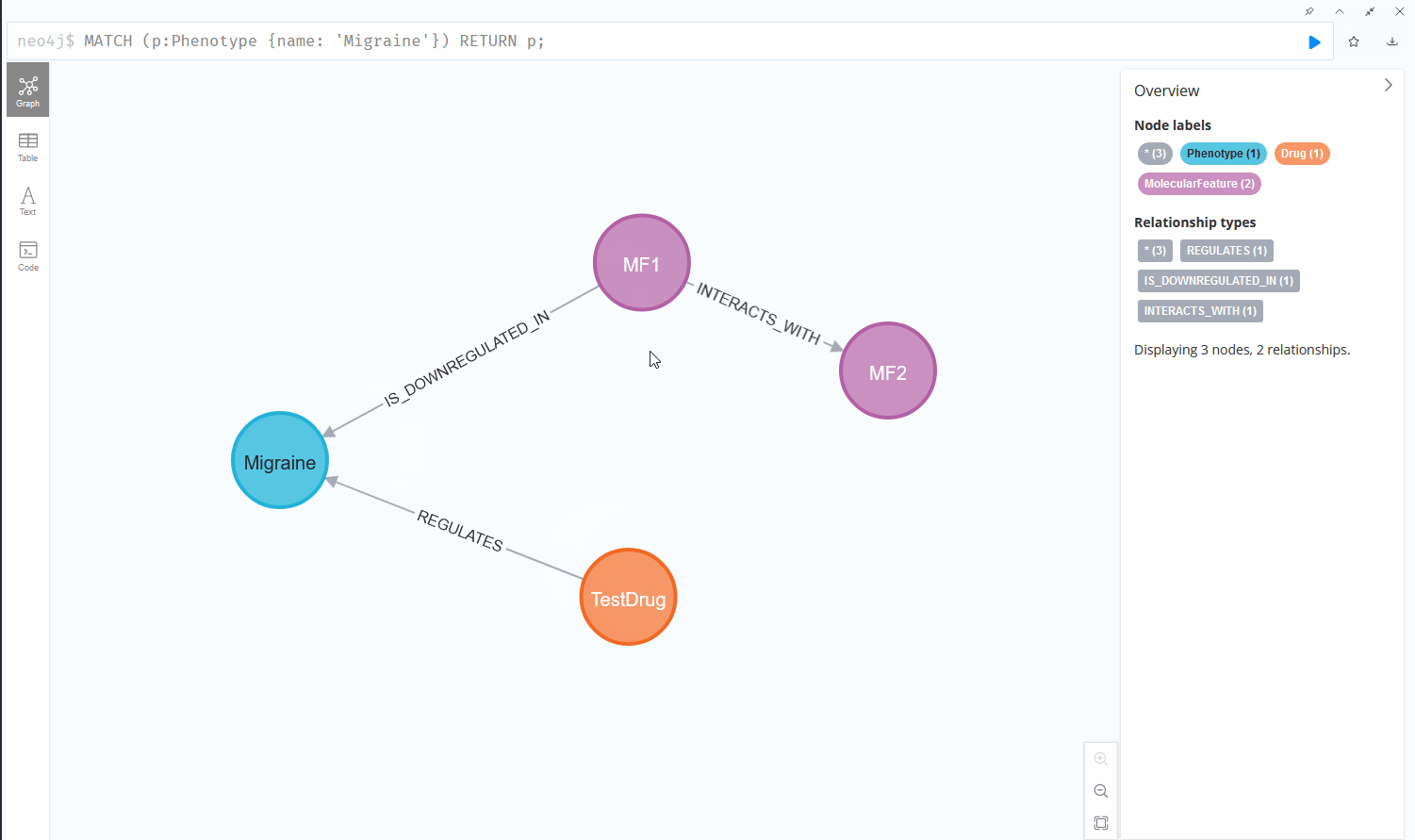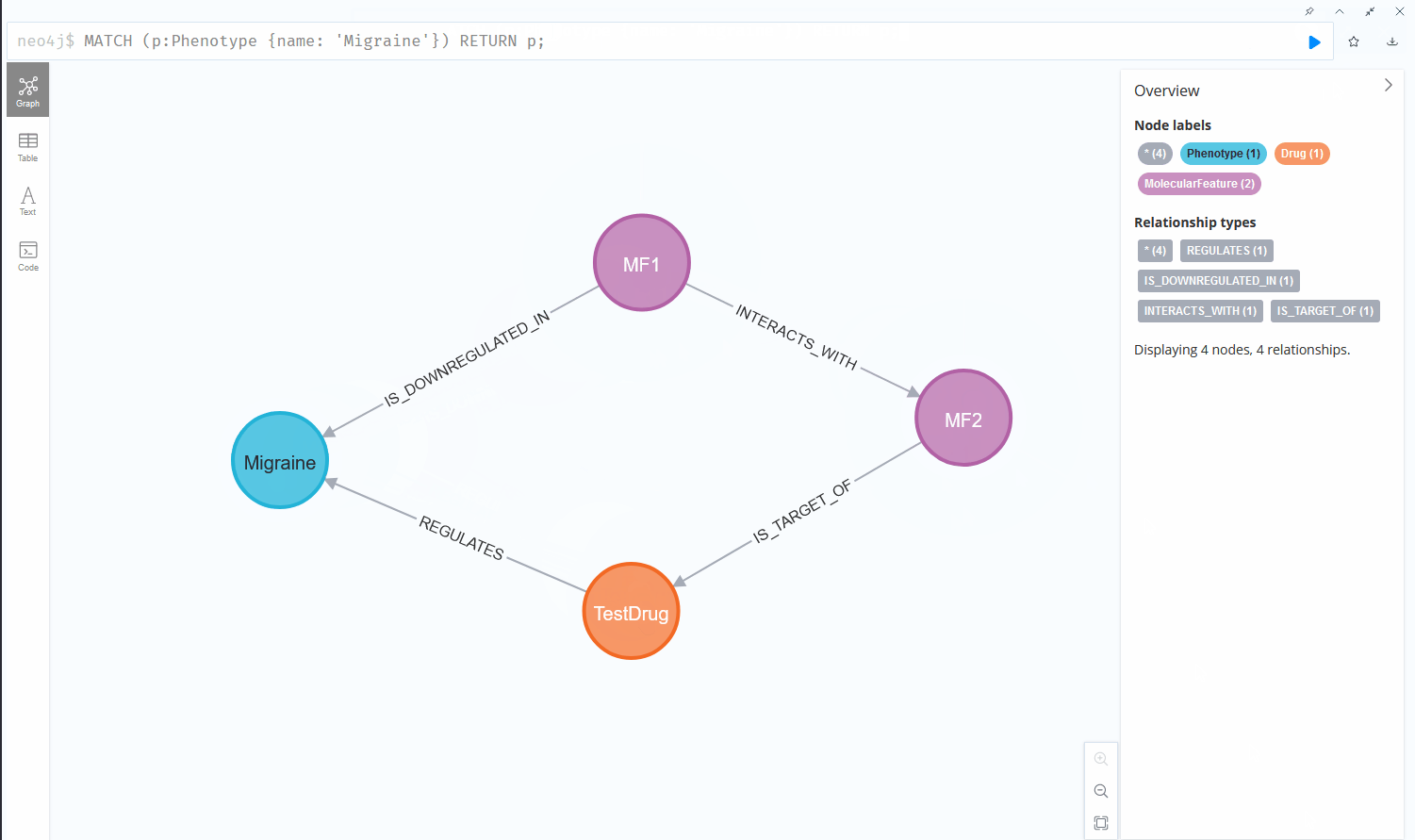
The same information represented as a graph maintaining the same degree of information by encoding details of the table into edges and properties can be displayed like this:
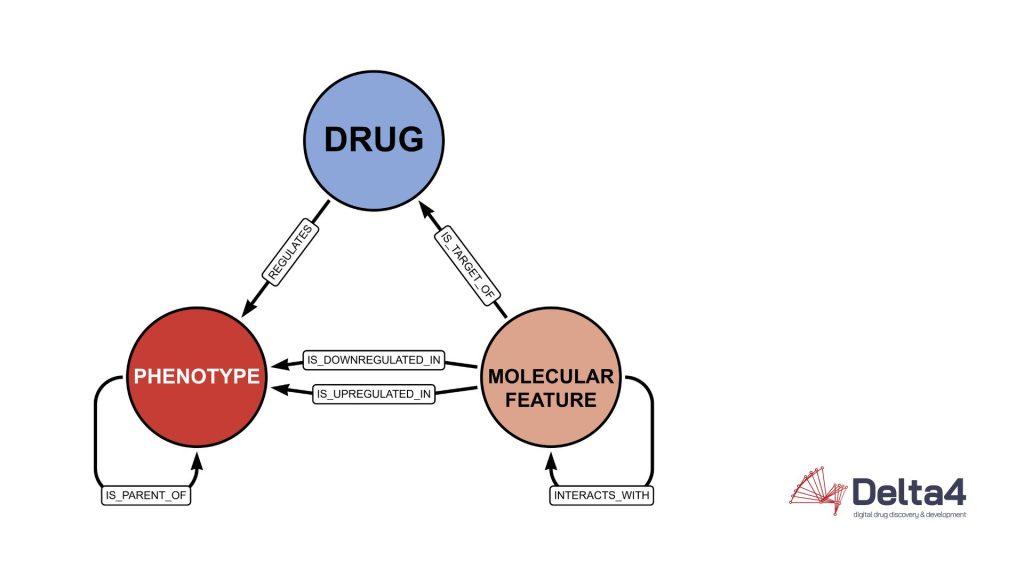
The graph structure proves its worth in simplicity and readability.
To retrieve data from the databases, for example, to find all drugs that play a role in migraine treatment, the graph database uses the following syntax:
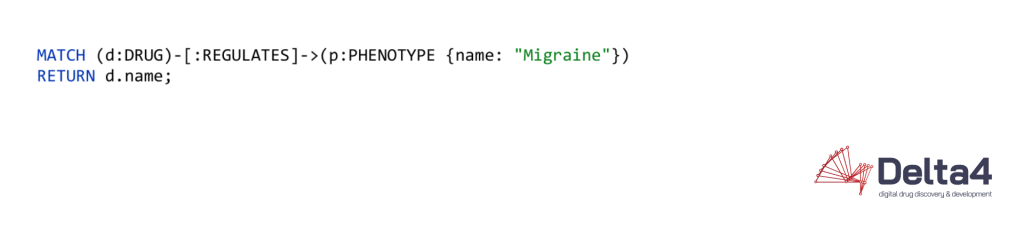
The equivalent in a RDBMS looks like this:

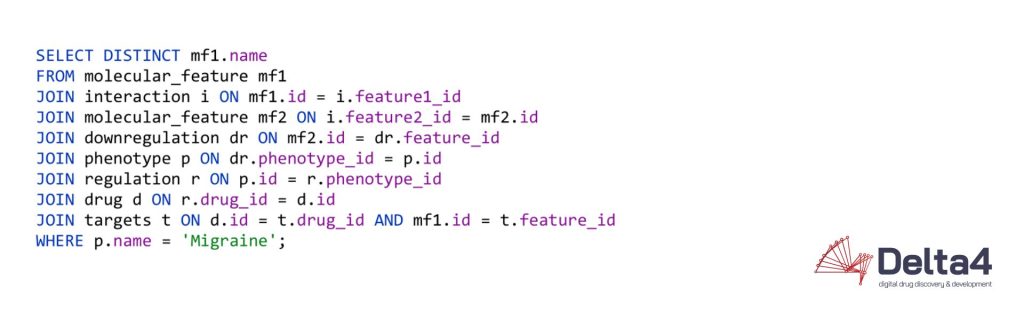
Now if we want to find all molecular features that interact with another molecular feature that are furthermore downregulated in a certain phenotype, the query quickly becomes more complex and the usability of both systems starts to diverge:

This chain of relationships might seem rather intuitive in cypher, the query language used by Neo4j, one of the more prominent graph database vendors. We go from node to node by specifying the relationships we want to investigate, written in a syntax that reads like a logical sequence of conditions. Querying the same data could look like this in a Structured Query Language (SQL) statement, used by RDBMS:
Network Algorithms and Machine Learning
Simplicity and readability are not the only benefits of graph databases. They enable network algorithms, such as calculations of similarities of nodes, clustering algorithms, finding the shortest path in a network, generation of embeddings that enable machine learning and AI methods, and many more. To broaden the usability of the system, many solutions come with integrated web interfaces that allow exploration of the data in an intuitive way, where one can explore the neighborhood of a network, construct and visualize queries that are easy to grasp.
Delta4’s Approach to Drug Discovery
At Delta4, we firmly believe in the importance and efficacy of both RDBMS and graph databases. Each system, unique in its capabilities, is instrumental in constructing robust, flexible pipelines tailored for the intricate processes involved in digital drug discovery.
Hyper-C Platform
Our Hyper-C platform capitalizes on the proven efficiency and reliability of RDBMS to ingest, harmonize, and manage a broad spectrum of data sources. Strengthened by our cumulative expertise, this facilitates the creation of a solid, unwavering data layer.
Hyper-KG Platform
Augmenting our architecture with graph databases, our cutting-edge product Hyper-KG is designed to leverage the interconnectedness of data, granting us the flexibility to navigate complex relationships, unravel hidden patterns, and empower AI tasks efficiently.
Conclusion
The parallel utilization of both RDBMS and graph databases catalyzes our mission, setting new benchmarks in the realm of digital drug discovery. By harnessing the power of knowledge graphs, Delta4 is paving the way for groundbreaking advancements in drug discovery, ultimately aiming to improve patient outcomes and transform the future of healthcare.


